Bioinformatics and Evolution of Cell Walls in Fungi
Bioinformatics and
protein structure:
We use bioinformatic tools to compare properties of protein
sequences. The amino acid sequences tell what types of domains are present, and
predict how they fold. The homology modeling shown on the agglutinins page is one example of this
approach. Another is our current
project to model the Threonine-rich glycosylated repeats in Als proteins.
Comparative studies are exemplified in an illustration of the sequences of fungal agglutinins and adhesins. The figure below was generated with the program DRAWHCA available through the EXPASY website. The figure illustrates repeat sequences (as boxed regions with recurring patterns); Cysteine residues as triangles placed beneath the sequences; N-glycosylation sites as hexagons above the sequences; and the frequency of threonine residues as dotted and solid lines beneath the sequences.
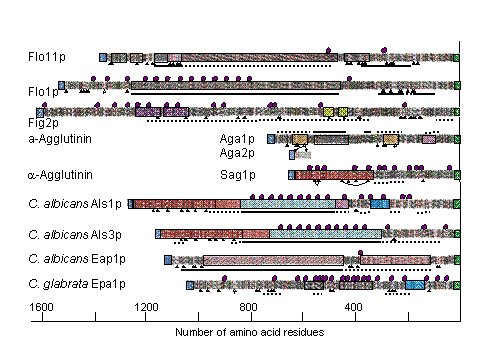
Evolution of Fungal Cell Walls
In collaboration with Dr. Wei-Gang Qiu and Dr. Susan Epstein of Hunter College, Juan Coronado has devised a new tool for comparison of serine- and threonine-rich sequences like those in cell wall proteins. We have used Juan’s method to study the evolution of cell wall proteins, and to study how fungal cell walls came to be developed, considering that the animals are the eukaryote kingdom most-closely related to fungi.
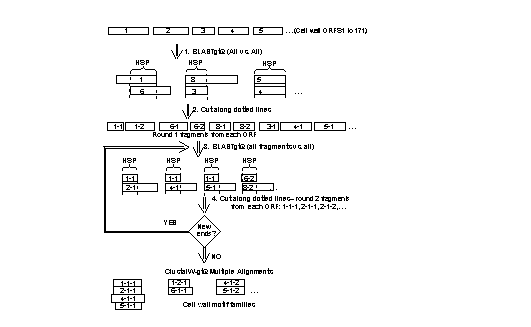
Procedure
for identifying repeated motifs in cell wall proteins from the MATCH paper
cited below
Coronado,
J.E., O. Attie, S.L.Epstein, W.-G Qiu, and P.N.Lipke. 2006.
Composition-modified matrices improve identification
of homologs of S. cerevisiae low-complexity
glycoproteins. Euk. Cell 5: 628-637.
Romov, P.A., F. Li, P.N. Lipke, S.L. Epstein, and W.G.
Qiu, W.-G. 2006. Comparative genomics reveals
long, evolutionarily-conserved, low-complexity islands in yeast proteins. J. Mol.
Evolution 63: 415-425.
Coronado, J.E., W.-G. Qiu, S.L. Epstein, and P.N. Lipke. 2007. Discovery of repeated sequence motifs in yeast cell wall proteins. MATCH: DIMACS 2006 in press